alpha and beta diversity maps
Jean-Baptiste Féret, Florian de Boissieu
2025-01-09
Source:vignettes/biodivMapR_06.Rmd
biodivMapR_06.Rmd
and
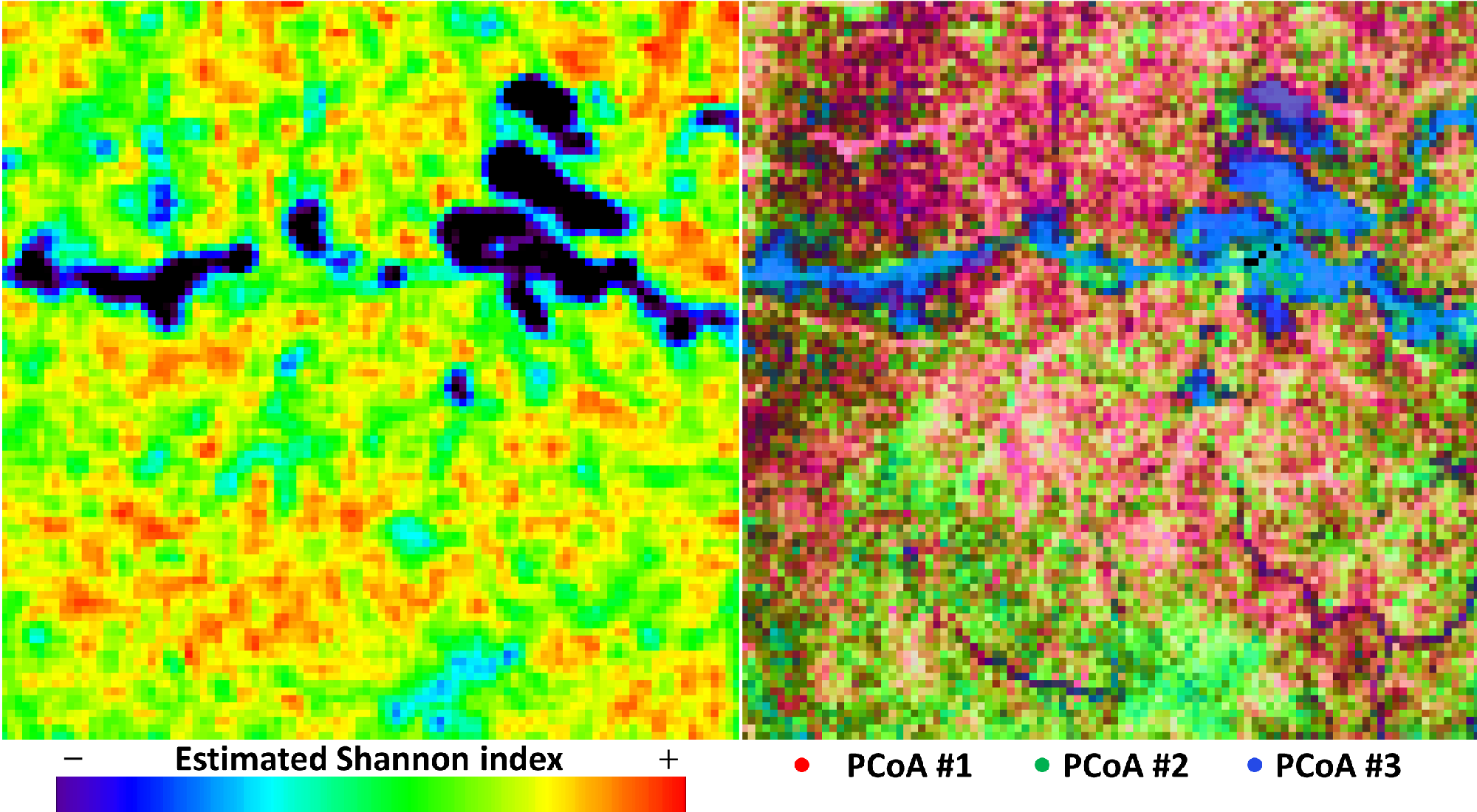
diversity maps are based on the SpectralSpecies raster.
The code to compute and diversity maps from this file is as follows:
print("MAP ALPHA DIVERSITY")
# Index.Alpha = c('Shannon','Simpson')
Index_Alpha <- c('Shannon')
map_alpha_div(Input_Image_File = Input_Image_File,
Output_Dir = Output_Dir,
TypePCA = TypePCA,
window_size = window_size,
nbCPU = nbCPU,
MaxRAM = MaxRAM,
Index_Alpha = Index_Alpha,
nbclusters = nbclusters)
print("MAP BETA DIVERSITY")
map_beta_div(Input_Image_File = Input_Image_File,
Output_Dir = Output_Dir,
TypePCA = TypePCA,
window_size = window_size,
nbCPU = nbCPU,
MaxRAM = MaxRAM,
nbclusters = nbclusters)and diversity maps are then stored in raster files located here:
RESULTS/S2A_T33NUD_20180104_Subset/SPCA/ALPHA
and here:
RESULTS/S2A_T33NUD_20180104_Subset/SPCA/BETA
Different types of rasters can be produced (full resolution or
resolution defined with window_size). Users are invited to
refer to the documentation for more options.
Here, processing our example leads to the following and diversity maps:

RGB representation of Sentinel-2 image.
Alternative diversity indices corresponding to functional diversity indices can also be computed. These do not require computation of the spectral species. They only require the PCA file or any stacked file.
The computation of teh functional diversity metrics is introduced in the next step.